P2N Data model¶
Generalities¶
The following scheme, inspired by [Abbas et al] presents the general organisation of the Patent2Net script suite. The differents scripts of Patent2Net adresses the pre-processing and processing steps. Post processing concerns human interactions with the tool.
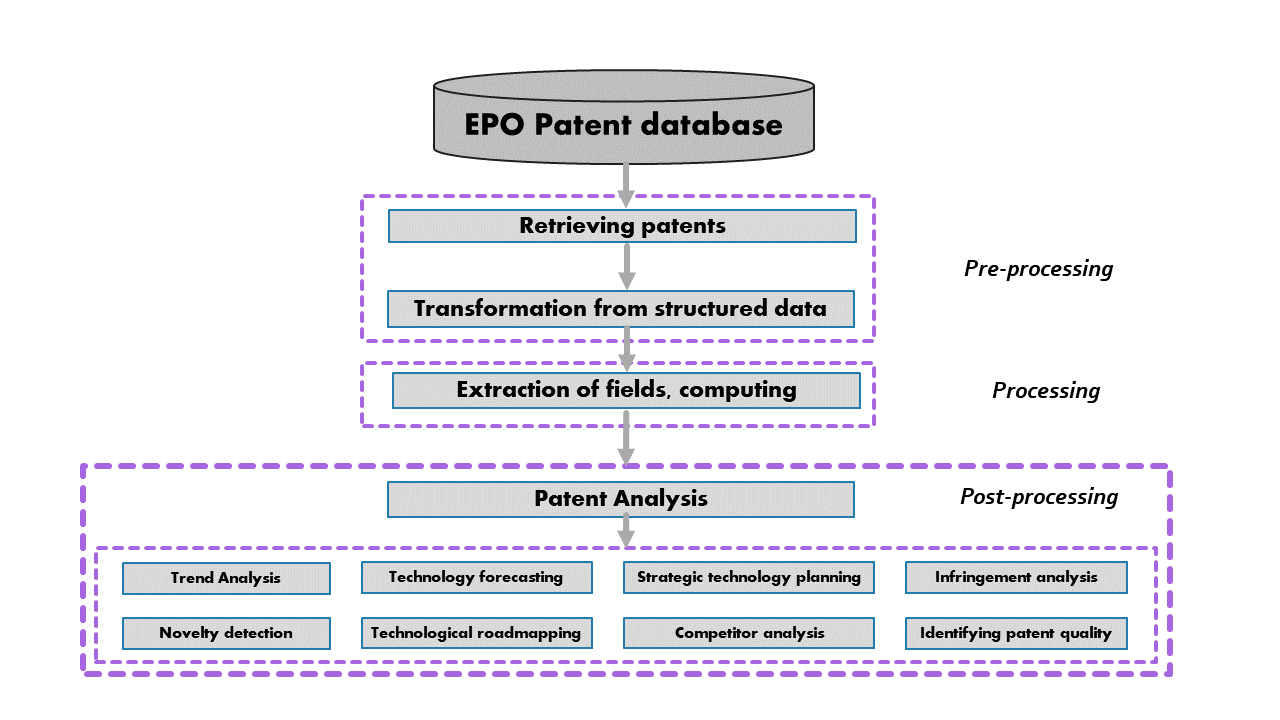
Script philosophy¶
The Generic philophy for a script is to follow the following procedure: - open a request file (requete.cql) and set the configuration
read the CQL request
memorize the Data directory (../DATA/Datadir) where all the stuff concerning gatherers and processors will be stocked
set up boolean switches (to do or not the job)
if the according boolean for the script is set to True, go ahead
Script organisation¶
The following scheme, presents the general organisation of the Patent2Net scripts in two categories: main and libraries.
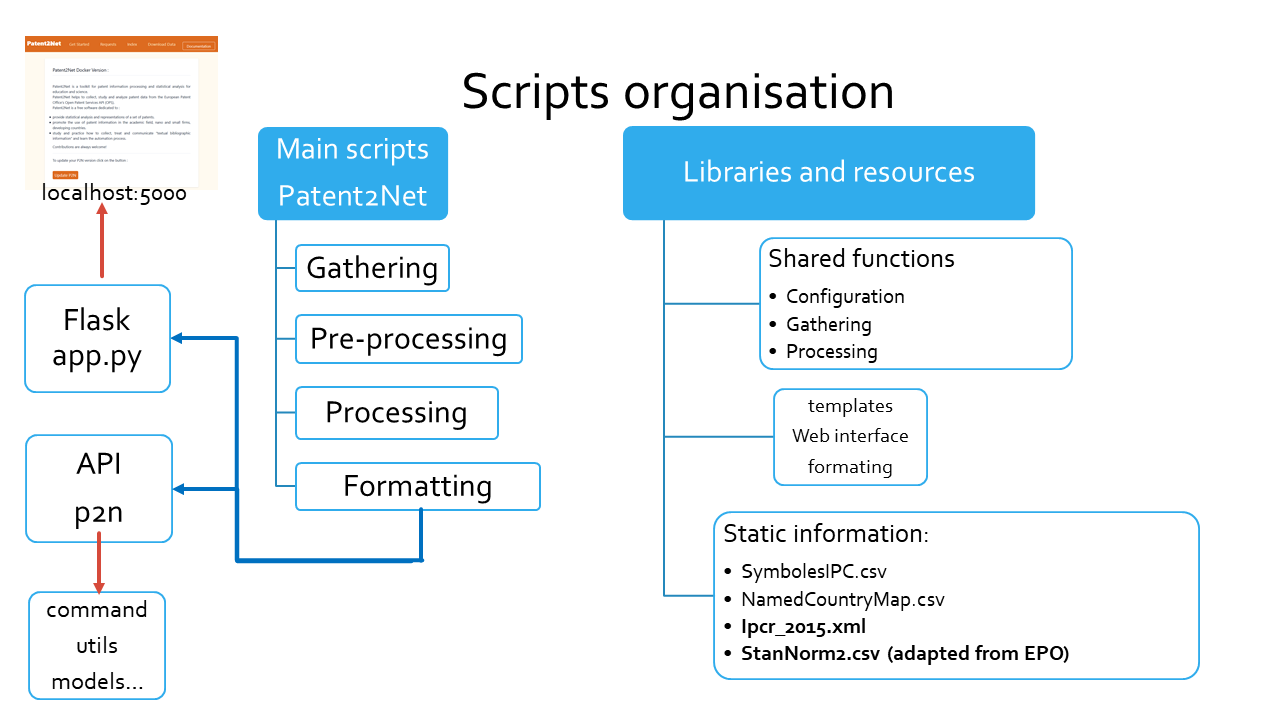
Main scripts¶
These scripts are separated in the four categories appearing on previous figure. They follow a generic named convention.
Gatherers¶
a gatherer starts with OPS-Gather. They use the wonderfull [epo-ops] library and they stand for:
- patent gathering. OPS-GatherPatentsv2.py is the entry gatherer. It creates:
the list of patent according to the cql request retreived to the [API]. This list is stocked in ../DATA/Datadir/PatentLists in pickeled dump format of the dictionnary returned by the API [1].
It gathers then the bibliographic data: the complete set of patent metadata for each entry in previous list. This data is stocked in ../DATA/Datadir/PatentBiblios in pickeled dump format of a list of dictionnary extracted from data.
patent familly gathering: for each patent in PatentBiblios, retreive its familly set of patent and add it.
patent images gathering: for each patent in PatentBiblios, retreive its media stuff and store it in ../DATA/Datadir/PatentImages
patent content gathering: for each patent in PatentBiblios, retreive its [abstract, Claims and Descriptions] if present in the databas. Store it in ../DATA/Datadir/PatentContents/[theGoodPart]
Pre-Processors¶
Data from the databasse comes most often dirty. Two scripts, added recently, prepares and clean it in two ways: 1. a filtering process to exclude from patent list equivalents patents. Only the oldest one in the dataset is kept. This process avoid biaises in text formating and many representations. Patent label excluded will be present in equivalents networks or families datasets.
Names of applicants and inventor aren’t normalized (swap of first name and name, very huge variations of applicant names). I add a preprocess to try to normalize case : leveinsthein distance and a huge database offered by the [EPO].
Processors¶
a processor script. This category is divided mainly in two parts: Fusionner that adresses the production of textual data [XML], [TXT] or Formatters that prepares data for visualisation tools (json files):
Fusion (Images, Carrot2, IRAMuTeQ) respectively prepares data for images visualisation, Carrot2 files in stadalone mode or via the Elastic Search plugin, and IRAMuTeQ formated textual files [IRAMuTeQ].
FormateExport* scripts prepares data for DataTables [Datatable], PivotTables [PivotTable.js], D3Js maps [D3plus], and Zotero compatible files [Zotero].
P2N-Freeplane prepares data for a mind-map of IPC labels augmented with their WIPO’s definition.
P2N-Nets-new.py scripts prepares data for network representations:
the [GEXF-JS] used in the network HTML pages as an online network visualisation tool,
the [GEXF] files to use with [Gephi] or other network visualisation ans exploration tool,
the following networks are processed:
Collaboration networks: builds the representation of who works with.
Inventors collaborations: for each patent, each co-inventor is considered and attached to the first inventors. Data is stored in ../DATA/Datadir/GephiFiles/DataDir_Inventors.gexf”. [GEXF-JS] exports introduces a neatto [Graphviz] positioning of node, takes the same name followed by a “JS” i.e.: ../DATA/Datadir/GephiFiles/DataDir_InventorsJS.gexf. For this visualization tool, a javascript file is created from a model template and loaded with the adequate HTML model page;
_Applicant collaborations: for each patent, each co-applicant is considered and attached to the first one. Same convention of naming and JS format files;
_Applicant and patents net: for each patent, each applicant is considered and attached to the label;
_Applicant, inventors and patents net: for each patent, each applicant is considered and attached to the label and so on for the author.
Bibliographic networks: builds the representation of where do this technology comes from or it is used.
patents and their Equivalents: for each patent, each equivalent label is attached to the main label (the oldest in the dataset, equivalents from this one are excluded by the filtering process);
patent and the References they cite: for each patent, each reference (label or non patent reference) is attached to its label
patent Citations: for each patent, each citation it receives from labels in the patent database is attached to its label.
Technologic networks: builds the representations of the mixed technology in application, and the expertises applicants or inventors are attached to:
Technology crossing: the DataDir_CrossTech networks represents for each patent and each International Patent Classification (IPC), the co-occurence of each technology code (from 4 digits to 11);
Authors and technology: the DataDir_Inventors_CrossTech represents for each inventor name a link to each IPC code owned patents declares;
Applicants and technologies: the DataDir_Applicants_CrossTech represents for each applicant name a link to each IPC code owned patents declares.
Note
Each node in the differents networks file is qualified with usefull caracterics acccording to the node category: families lenght, IPC force and diversity, or number of patent owned. Gephi help in computing several network well known caracteristics and visualization of them.
Visualization scripts¶
Caution
theses script are in beta version (more than others I mean)
Trizifyer¶
Clusterer¶
Double clusterisation process.
Indexer¶
Caution
this script is also in beta version.
P2N-Indexer is intended to feed Elastic Search with the documents containing the text of patent data gathered. Results of this new feature are presented thought the clusterer carrot2 elasicsearch plugin. Page presentation expects some new features coming soon.
Interface script¶
The script interface2 prepares the /DATA/DataDir/DataDir.html file using the config file and the template to present the whole processed files.
Note
The image below represents the general directory structure and schematic data flows in Patent2Net.
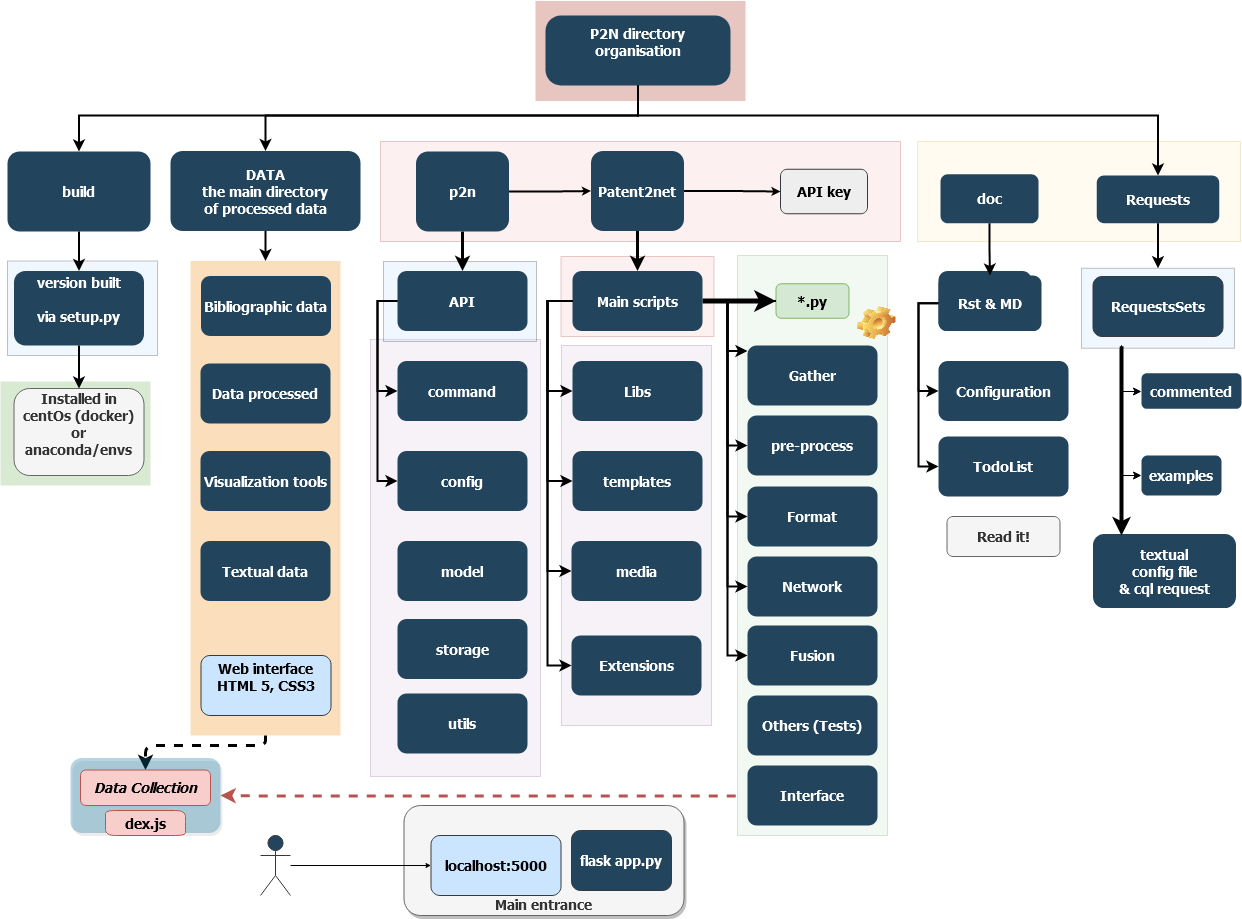
Other cool stuff¶
Caution
the feautures of the scripts below are only accessible via bash mode assuming you have some computer skills.
import label list from csv file in order to produce a PatentList compatible with OPS-Gather*.
Request splitters: AutomRequestSplitter* allow to split a request in time and/or over the IPC codes to produce requests that do not exceed the [API] 2000’ limit
footnotes